2023-11-15
Review: Overview of git and GitHub
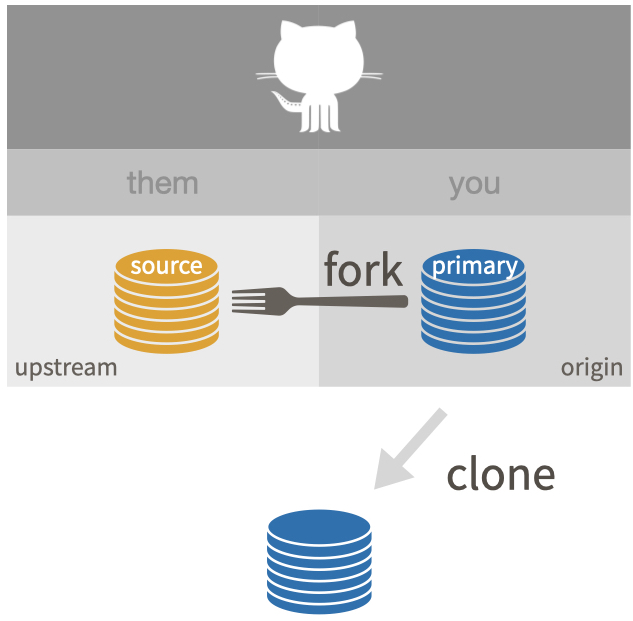
Forking and cloning
Branching, merging, and GitHub Flow

Activity: Providing feedback on README.md files
- Exchange your GitHub usernames and project repo names with someone else
- You’ll each provide feedback on the other’s
README.mdfile - Go to their repo on GitHub, look over their
README.mdand write down questions and constructive feedback - On GitHub, click “Issues”, then “New issue”
- Put in your feedback with a title
Assigned readings jigsaw
- 05 mins: With expert group discuss main points of your reading
- 15 mins: As the expert, share what you learned
- 15 mins: Class discussion
Break time!

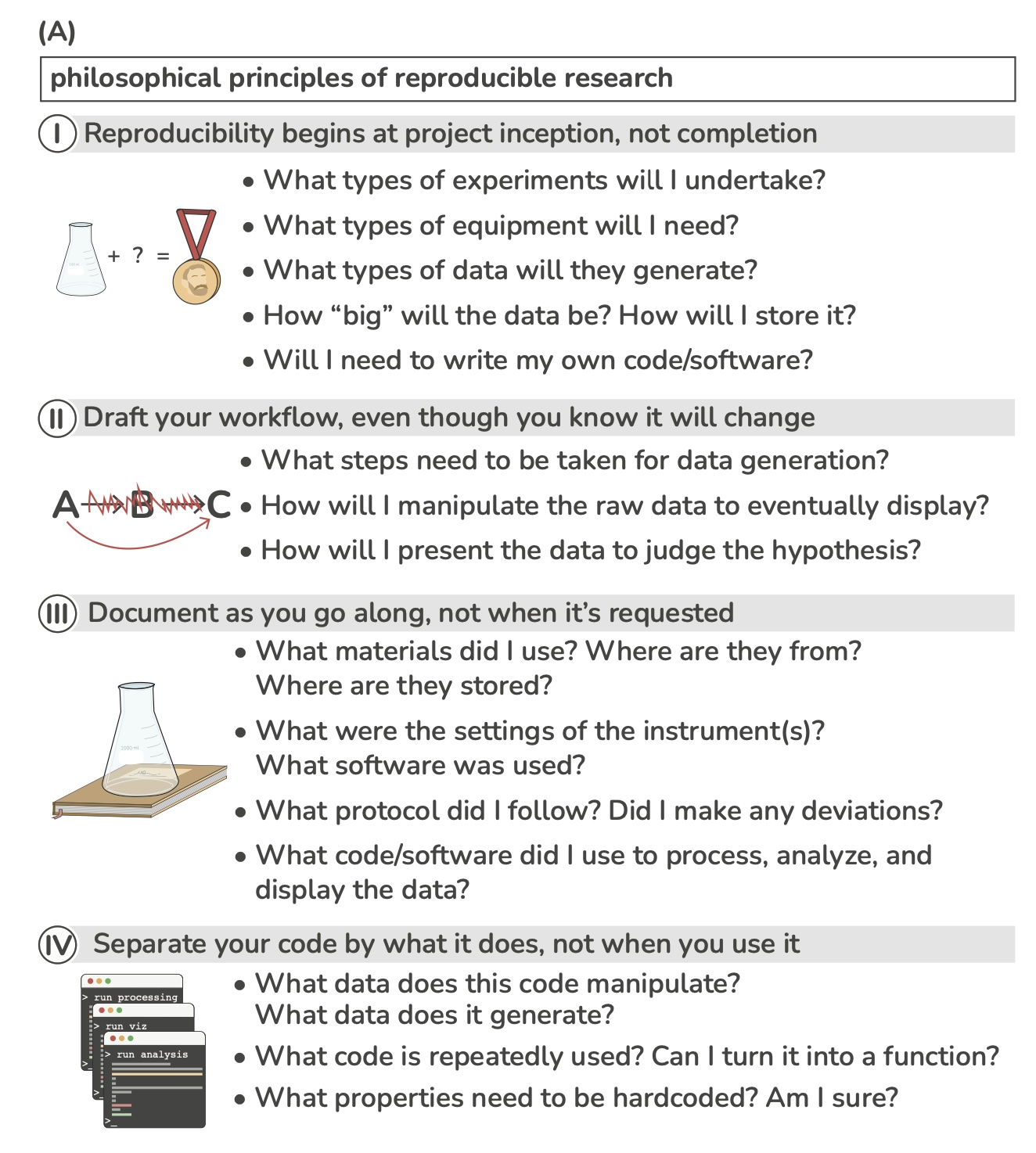
Why do file management and workflow matter?
If you quit today and dropped your project, could a someone pick take over your project and work with your data without talking to you?
Do you think you could come back to this project in 15 years and reproduce your analysis?
What makes up a “good analysis workflow”
- Analysis starts from copies of the raw data
- Deny your permission to write on the raw data files
- Do all data wrangling with scripts - no point-and-click
- Separate your scripts into what they do
- Loading, cleaning, and wrangling the data
- Data analysis
- Create visualizations, tables, statistics
- Intermediary files are kept seperate from the raw data
- WET vs. DRY
- “Write everything twice”
- “Don’t repeat yourself”
Draft your workflow
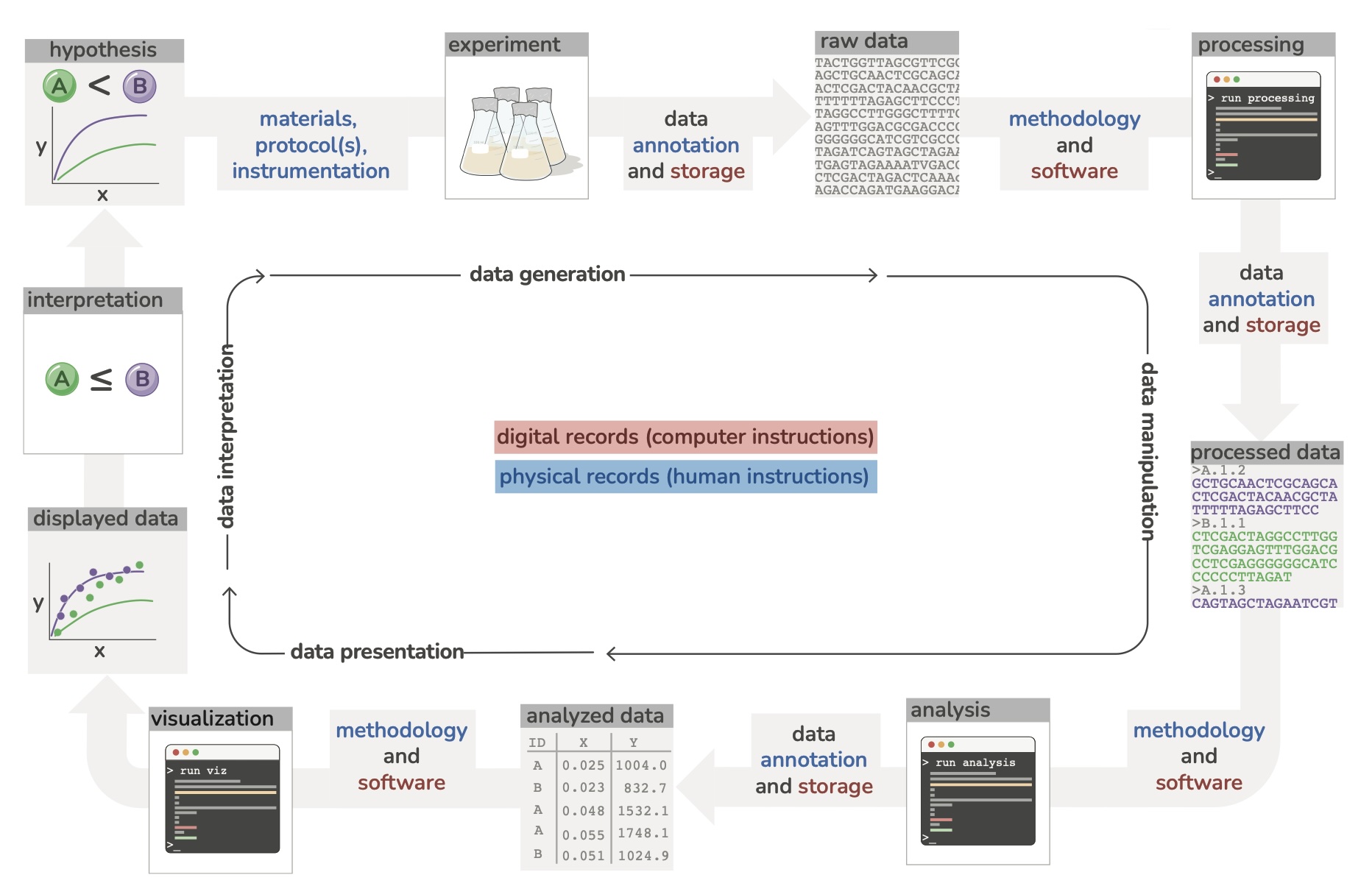
Draft your workflow
- This is a living structure
- How is the data generated?
- Primary vs. secondary data
- Type of data
*.xlsx,*.csv,*.fastq,*.gff,*.tsv, images, etc.
- Track materials, instrument(s), software, etc.
- How big is the data? Where will it be stored?
- How is the raw data manipulated?
- Where is the protocol and any changes made documented?
- How will the data be presented/visualized?
Answer these questions for your project
Example of a basic structure for a project directoryList from https://www.britishecologicalsociety.org/wp-content/uploads/2019/06/BES-Guide-Reproducible-Code-2019.pdf
The data folder contains all input data (and metadat) used in the analysis.
The doc folder contains the manuscript
The figs directory contains figures generated by the analysis
The output folder contains any type of intermediate or output files (e.g. simulation outputs, models, processed datasets, etc.). You might separate this and also have a cleaned-data folder.
The R directory contains R scripts with function definitions.
The reports folder contains RMarkdown files that document the analysis or report on results.
The scripts that actually do things are stored in the root directory, but if your project has many scripts, you might want to organise them in a directory of their own.
Example project directory
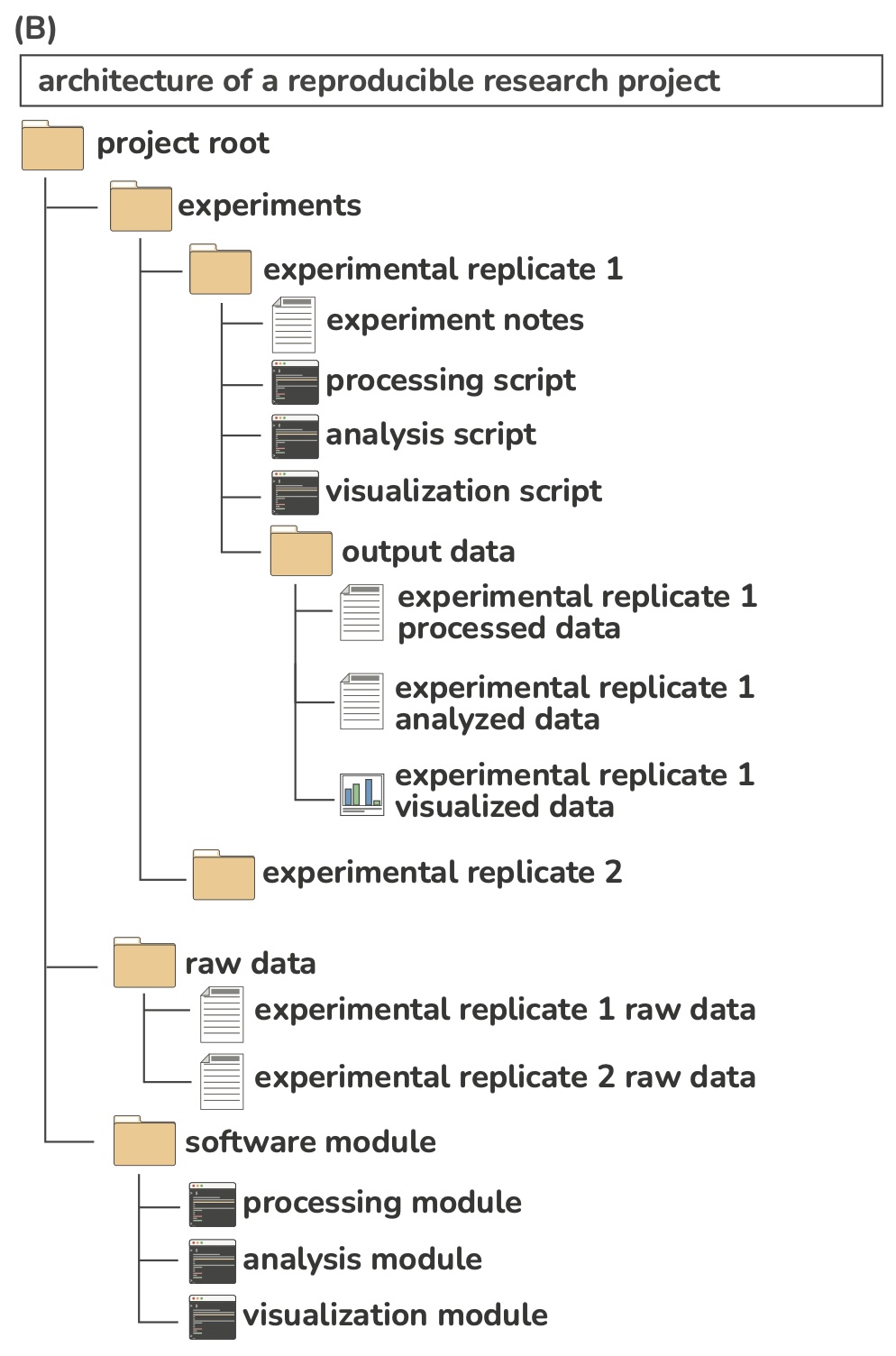
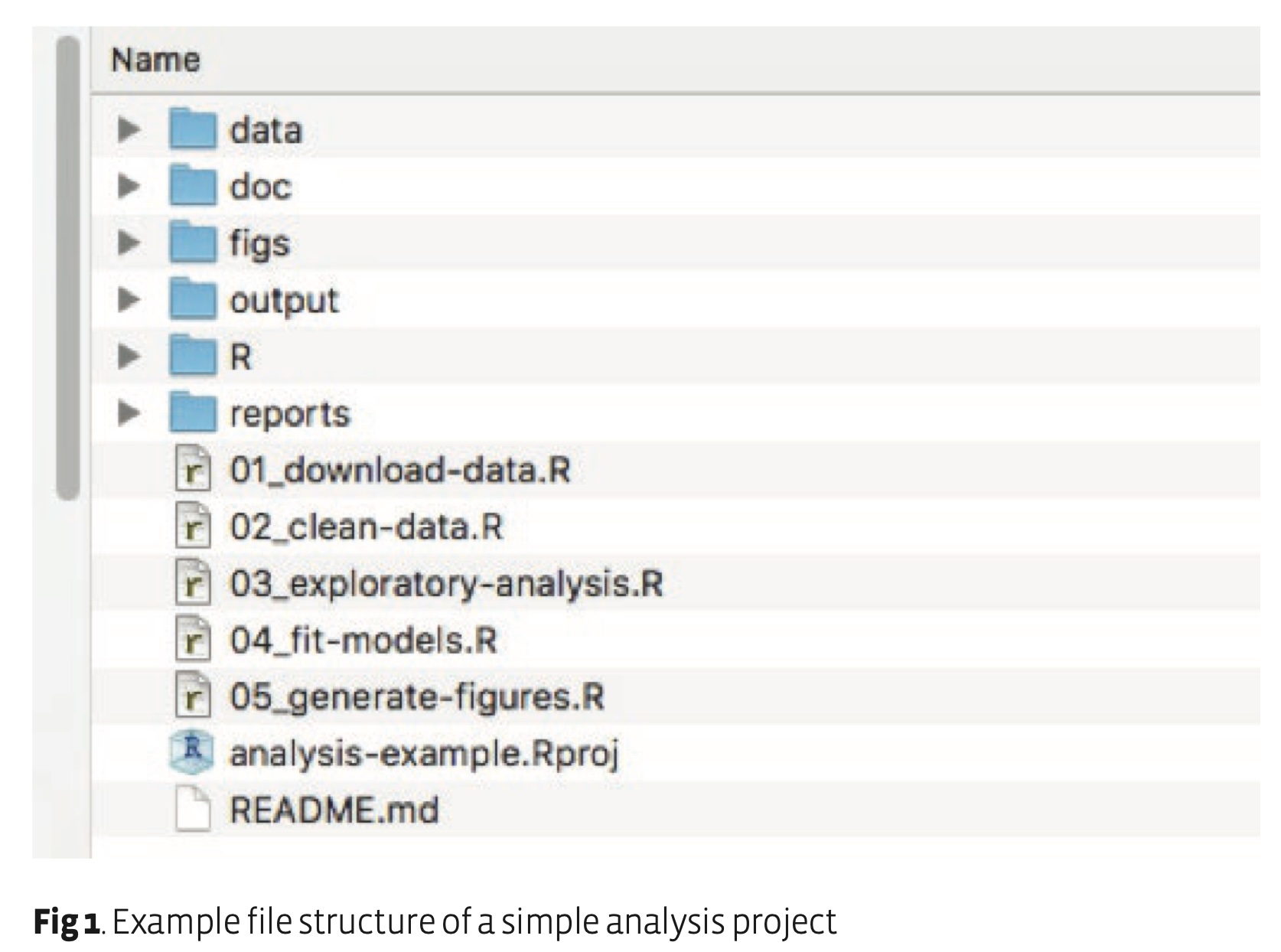
Folder structure
Have a consistent naming scheme
Arrange folders in hierarchical structure
Have a README file that describes the project as well as a basic tour of your folder structure
Include README files in subfolders for files that aren’t described or commented easily
- Include description of file contents, content source(s), relevant papers, etc.
Include an appropriate license
Seperate in progress and completed work
Keep track of ideas, notes, discussions and next steps with GitHub Issues
File namingList from https://www.britishecologicalsociety.org/wp-content/uploads/2019/06/BES-Guide-Reproducible-Code-2019.pdf
- Machine readable
- Avoid spaces, punctuation, accented characters and case sensitivity.
- More specifically, stick to “a-zA-Z0- 9_” characters.
- Use periods/full stops for file type only (i.e.
*.csv). - Use delimiters to separate and make important metadata information (for example parameter values used in an analysis) retrievable further down the line. Use delimiters consistently, i.e. “_” to separate metadata to be extracted as strings later on and “-” instead of spaces or vice versa but do not mix. This makes names easy to match and search programmatically and easy to analyse.
- Avoid spaces, punctuation, accented characters and case sensitivity.
- Human readable
- Ensure file names also include informative description of file contents.
- Adapt the concept of the slug to link outputs with the scripts in which they are generated.
File namingList from https://www.britishecologicalsociety.org/wp-content/uploads/2019/06/BES-Guide-Reproducible-Code-2019.pdf
- Easy to order by default
- Starting file names with a number helps.
- For data, this might be a date allowing chronological ordering.
- Make sure to use ISO 8601 format (YYYY-MM-DD) to avoid confusion between differing local dating conventions.
- For scripts, you could use a number indicating the position of the scripts in the analysis sequence e.g. 01_download-data.R
- Make sure you left-pad single digit numbers with a zero or you will end up with this:
- 10_final-figs-for-publication.R
- 1_data-cleaning.R
- 2_fit-model.R
Using Griffin Chure’s workflow template
Navigate to https://github.com/gchure/reproducible_research
Click on green “Use this template”
Select “Create a new repository”
Clone the new repository to your local machine
Copy and paste the folders into your project repository
Play with the structure, folders, naming to fit your own project
Conclusion
File management incorporates naming and structure
Workflow helps you organize your projects and make it easier for others to reproduce your work